Team
Coordination

DR, CNRS
Cell-ID Scientific Director

DR, CNRS
Scientific leader of PC1

DR, CNRS
Scientific co-leader of PC1


DR, CNRS
Scientific leader of PC2

DR, Inserm
Scientific co-leader of PC2

DR, CNRS
Scientific leader of PC3 Data

PR, Mines
Scientific co-leader of PC3 Data

DR, Inserm
Scientific leader of PC3 Med

DR, CNRS
Scientific leader of PC3 Med


DR, CNRS
Scientific Leader of PC4

DR, CNRS
Scientific co-leader of PC4

PR, Mines
Scientific co-leader of PC4
Support Team


(CNRS)
Governance, Cell Context & Cell Exp

(Inserm)
Cell Next

(Curie)
Data Med


(Curie)


(CNRS)


(CNRS)

(Inserm)
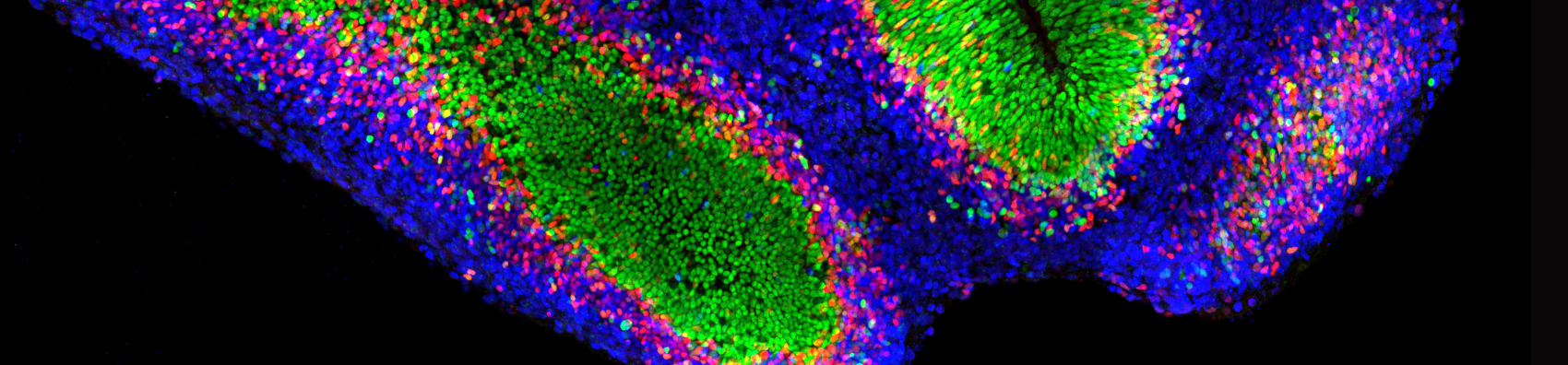
Cell Context | Cells in space and time

Research: Epigenetics, Chromatin biology, 3D genome organization, epigenetic inheritance and memory, Chromatin state transitions during differentiation and senescence
Methods: Chromosome Conformation Capture (3C, 4C, 5C, Hi-C, Capture-Hi-C, ChIA-PET, HiChIP), Chromatin Immunoprecipitation (ChIP, ChIP-seq), RNA sequencing (bulk & single-cell), DNA methylation profiling, Drosophila genetics, Protein–DNA mapping, Bioinformatics and Statistical modeling of gene regulation, Microscopy analysis
Technologies: Next-Generation Sequencing (NGS) platforms, Hi-C and related chromatin conformation technologies, ChIP-seq and CUT&RUN / CUT&Tag platforms, Confocal and live-cell microscopy, Super-resolution microscopy (STORM, SIM)

Research: Single-cell genomics and transcriptomic, Lung tissue homeostasis and repair mechanisms, Systems biology of lung diseases
Methods: Single-cell RNA sequencing and transcriptomics, High-throughput sequencing technologies, Multi-omics integration, Data mining and machine learning approaches for genomic data, Microfluidic devices for cell indexing and sequencing
Technologies: Highthroughput sequecing technolofies (nanopore and illumina), Fluidigm C1 microfluidic device for single-cell analysis, High-content and adaptive optics microscopy, Computational tools for single-cell and multi-omics data analysis

Research: Proteomics of chromatin and nuclear dynamics, DNA damage response and repair mechanisms, post-translational modifications regulating genome stability, integration of proteomic and transcriptomic data to study cellular stress responses.
Methods: Quantitative and functional proteomics (LC–MS/MS, SILAC, label-free), Chromatin Immunoprecipitation (ChIP) and complex purification, RNA sequencing and integrative omics, Fluorescence and confocal microscopy for subcellular protein localization.
Technologies: Advanced mass spectrometry platforms, High-throughput proteomics and data analysis pipelines, Bioinformatics tools for protein network modeling and systems biology, Integrated imaging–proteomics workflows for spatial and molecular profiling.

Research: Epigenetic plasticity in cancer; single-cell epigenomics; early tumor evolution and therapy resistance; non-genetic mechanisms of breast cancer progression; chromatin dynamics and transcriptional regulation in response to treatment.
Methods: Single-cell chromatin immunoprecipitation sequencing (scChIP-seq); high-throughput droplet-based epigenomic profiling; RNA sequencing; spatial transcriptomics; computational analysis of epigenomic landscapes; development of interactive tools for data interpretation.
Technologies: Single-cell and spatial omics platforms; high-resolution sequencing technologies; bioinformatics pipelines for single-cell epigenomic data; advanced imaging systems for chromatin visualization; custom software for epigenomic data analysis and visualization.

Research: Chromatin structure & 3D genome organization, Transcription regulation, Single-cell variability in gene expression and chromatin folding, DNA segregation complexes
Methods: Hi‑M: multiplexed DNA/RNA imaging (spatial genomics), Super-resolution microscopy (STORM, multifocus microscopy), Single-molecule localization microscopy (SMLM), Live imaging, Magnetic tweezers, Microfluidics
Technologies: Adaptive optics, Spatial genomics/transcriptomics pipelines, Long-range chromatin mapping, chromatin loops visualization, Multiplex imaging software, Data analysis platforms, chromatin folding modeling

Research: Gene expression mechanisms, RNA metabolism, Bimolecular condensates, Retroviral RNA dynamics, Transcriptional regulation
Methods: Single-molecule imaging, RNA tagging and visualization, Protein-RNA interaction analysis
Technologies: MS2-based RNA tagging systems,High-resolution imaging systems for observing cellular processes,Live-cell imaging setups: Equipment designed for imaging living cells over time, Image analysis software

Research: DNA double-strand break repair, chromatin remodeling and genome stability, spatial and temporal organization of DNA damage response, interplay between transcription and repair, epigenetic regulation of genome maintenance
Methods: Inducible site-specific DNA break systems (AsiSI, CRISPR-Cas9), Chromatin Immunoprecipitation (ChIP-seq, ChIP-exo), RNA sequencing and nascent RNA mapping, Hi-C and related chromosome conformation capture assays, Super-resolution and live-cell microscopy to visualize DNA repair foci.
Technologies: Next-generation sequencing and integrative omics platforms, Imaging systems for real-time chromatin dynamics, Computational tools for mapping repair and transcription interactions, Automated pipelines for ChIP-seq and Hi-C data analysis.

Research: Advanced imaging techniques for single-cell analysis; integration of computational models with experimental data to understand chromatin dynamics; application of artificial intelligence to enhance bioimaging and drug discovery; investigation of the 3D organization of the genome and its impact on gene expression.
Methods: High-resolution microscopy including super-resolution and live-cell imaging; single-molecule RNA FISH (smFISH) and multiplexed RNA FISH; computational modeling of chromatin structure and dynamics; machine learning algorithms for image analysis and phenotypic screening.
Technologies: FISH-quant (MATLAB-based tool for RNA FISH quantification); QuickPALM (ImageJ plugin for super-resolution reconstruction); deep learning frameworks for image segmentation and analysis; high-throughput imaging systems for large-scale screening.

Research: Pediatric neuro-oncology, Tumor microenvironment, Tumorigenesis in cerebellar development, Tumor cell plasticity and resistance, Cancer cell heterogeneity, Pediatric cancer immunotherapy, Translational research in childhood oncology
Methods: Proteomics and mass spectrometry, Nanopore sequencing, Multi-omics approaches, ScRNA sequencing, In vivo mouse models, Tumor microenvironment analysis, High-throughput screening for therapeutic targets
Technologies: Advanced sequencing platforms, Mass spectrometry-based proteomics, Single-cell sequencing technologies, Multiplex immunofluorescence imaging, Data integration platforms for multi-omics data, Computational modeling of tumorigenesis

Research: Nuclear organization and 3D chromatin dynamics, Chromatin mechanics and transcription regulation, Physical principles of chromosome structure, Spatial transcriptomics and single-cell gene expression, Multiplexed RNA/DNA FISH, Tissue-level spatial organization of gene expression
Methods: Single-molecule microscopy, Micromanipulation of chromatin, Polymer modeling of chromosomes, Multiplexed RNA/DNA FISH, Xenium platform management (10X Genomics), Imaging-based spatial analysis of single cells
Technologies: Advanced microscopy, Magnetic manipulation tools for genomic loci, Computational modeling of chromatin dynamics, Xenium (10X Genomics) high-throughput spatial transcriptomics, Multiplexed FISH probe design, High-resolution imaging

Research: Quantitative RNA imaging; single-RNA regulation at different spatial scales; transcription & transcriptomics; developing workflows for spatial gene expression; combining microscopy with computational models and image analysis.
Methods: smFISH / sequential / multiplexed FISH; automated microscopy + image workflows; microfluidics; single-cell assays; image segmentation and bioimage analysis; open source software development for spatial transcriptomics.
Technologies: Advanced optical microscopy (widefield, confocal, live-cell imaging, SIM / SMLM etc.), robotics/automation for microscopy, platforms for high-throughput smFISH, software tools for segmentation and quantitative image‐based transcriptomics (e.g. RNA2seg, Comseg), data visualization and modeling.
Cell Exp | Experimental systems

Research: DNA double-strand break repair, chromatin remodeling and genome stability, spatial and temporal organization of DNA damage response, interplay between transcription and repair, epigenetic regulation of genome maintenance
Methods: Inducible site-specific DNA break systems (AsiSI, CRISPR-Cas9), Chromatin Immunoprecipitation (ChIP-seq, ChIP-exo), RNA sequencing and nascent RNA mapping, Hi-C and related chromosome conformation capture assays, Super-resolution and live-cell microscopy to visualize DNA repair foci.
Technologies: Next-generation sequencing and integrative omics platforms, Imaging systems for real-time chromatin dynamics, Computational tools for mapping repair and transcription interactions, Automated pipelines for ChIP-seq and Hi-C data analysis.

Research: Development, evolution and function of commissural systems in the nervous system; axon guidance and neuronal migration during brain development; mechanisms underlying brain connectivity; use of advanced imaging techniques to study neural circuits in vivo and human embryology.
Methods: Tissue clearing and light-sheet microscopy for 3D imaging of neural structures; whole-mount immunostaining to visualize axonal tracts and neuronal pathways; genetic manipulation in mouse models to study gene function in neural development; high-resolution imaging techniques to analyze brain architecture and connectivity.
Technologies: iDISCO protocol for clearing brain tissues and embryos; light-sheet microscopy systems for fast and high-resolution imaging; computational tools for 3D reconstruction and analysis of neural data; advanced imaging platforms for large-scale screening and data acquisition.

Research: Mechanisms of patterning and cell diversity during human neurodevelopment, morphogen signaling, chromatin dynamics, transcriptional regulation, neurodevelopmental and motor neuron diseases.
Methods: Generation and use of human pluripotent stem cell-derived models (organoids, specific cell types); single-cell transcriptomics; funtional genomics; histology; live-cell imaging.
Technologies: organoid culture and manipulation; genetic modifications of human pluripotent stem cells; Fluorescent in situ hybridization, high-throughput imaging; atac-seq; cut&tag and associated bioinformatics pipeline.

Research: Cell fate determination and tissue patterning during development; mechanisms driving neurogenesis and organ formation; spatial and temporal regulation of cell differentiation; modeling developmental processes in organoids.
Methods: Mouse genetics for in vivo functional analysis; live-cell imaging to track dynamic cellular behaviors; single-cell transcriptomics to profile gene expression; organoid culture to recapitulate tissue development.
Technologies: High-resolution imaging systems for dynamic cell analysis; bioinformatics pipelines for single-cell and spatial data interpretation; organoid culture platforms; computational modeling of developmental processes.

Research: Dynamics of neural stem cell division and differentiation in the developing human neocortex; mechanisms underlying the diversity of cortical cell types during embryonic development; evolutionary comparison of cortical development across primates; development of advanced live imaging techniques to track cell fate decisions in real time.
Methods: Long-term live-cell imaging of radial glial progenitor cells in cerebral organoids; semi-automated correlative imaging combining live and fixed tissue analysis; single-cell RNA sequencing to profile gene expression during neurogenesis; comparative analysis of human and non-human primate cortical organoids.
Technologies: High-resolution fluorescence microscopy systems for live imaging; organoid culture platforms for modeling human neurodevelopment; computational tools for image analysis and cell fate mapping; bioinformatics pipelines for integrating imaging data with transcriptomic profiles.
Data Med | Data processing for disease interception

Research : Chromatin organization, 3D genome architecture, gene regulation, epigenomics, polymer physics, statistical biophysics, physiological vs pathological states
Methods : Computational modelling, polymer physics, statistical mechanics, simulation of chromosome folding, quantitative and statistical tools, comparative genomics
Technologies : High‑throughput sequencing, imaging data, numerical simulations, computational pipelines, statistical inference, GPU or HPC for simulations, bioinformatics tools

Research : Integration of single-cell multi-omics across space and time, cellular trajectories, disease heterogeneity, regulatory mechanisms, translating high-dimensional omics into actionable biological insights
Methods : Multi-view learning, dimensionality reduction, network inference, graph‑convolutional networks, machine learning, benchmarking of multi-omics methods, computational modelling of cell states & transitions
Technologies : Single‑cell sequencing, multi‑omics, spatial omics, deep learning architectures, heterogeneous multilayer graphs, open‑source software for multi-modal data integration

Research : Spatial transcriptomics, computational pathology, high‑content screening, phenotypic screening, computational phenotyping, spatial modeling, computer vision
Methods : Deep learning, computer vision, artificial intelligence, machine learning, image segmentation, image classification, cross-modality prediction, predictive modeling, data integration.
Technologies : Neural networks, Deep Learning, Image Analysis, Foundation Models

Research : Bioinformatics, biostatistics, cancer genomics, high-throughput sequencing, somatic variant calling, data interoperability, personalized medicine, software engineering, high-performance computing
Methods : Development of biostatistical algorithms, genomic data analysis, data interoperability, high-performance computing, benchmarking of genomic tools, integration of multi-omics data
Technologies : High-throughput sequencing platforms, bioinformatics pipelines, patient-derived xenograft models, biomedical data interoperability systems, computational tools for genomic analysis

Research : Computational systems biology, tumorigenesis & tumor progression, cancer heterogeneity, multi‑omics data integration, precision medicine, prognostic & predictive modeling, signaling network models
Methods : Statistical analysis of high throughput biological data, independent component analysis, clustering and class discovery, machine learning, simulation, clinical, omics
Technologies : High‑throughput sequencing, bioinformatics pipelines, web‑based visualization tools, computational modeling & simulation, digital twins (in silico tumor modeling)

Research : Developmental biology, gene regulation, cellular differentiation, molecular pathways, stem cells, embryogenesis, genetic networks, signal transduction
Methods : Molecular cloning, gene expression analysis, CRISPR/Cas9 genome editing, microscopy, cell culture, RNA sequencing, functional assays, imaging techniques
Technologies : Next-generation sequencing, fluorescence microscopy, live-cell imaging, bioinformatics tools, flow cytometry, gene editing platforms, laboratory automation

Research : Computational biology, systems biology, cancer genomics, tumor microenvironment, epigenetics, data integration, machine learning, biological networks
Methods : Statistical modeling, bioinformatics analysis, high-throughput data analysis, algorithm development, data mining, network inference, predictive modeling
Technologies : Next-generation sequencing, computational pipelines, machine learning frameworks, data visualization tools, high-performance computing, databases, programming languages

Research : Pediatric brain tumors, H3K27‑altered DMG, epigenetic regulation, histone mutations, tumor heterogeneity, cancer genomics, molecular oncology, tumor classification, clinical translation
Methods : Next‑generation sequencing, whole‑exome, targeted sequencing, epigenomic profiling, genetic perturbations, tumor biopsy, in vivo models, stereotactic neurosurgery, in utero electroporation
Technologies : High-throughput sequencers, bioinformatics pipelines, epigenome analysis software, surgical stereotaxy tools, animal surgery suites, tissue culture instruments, transcriptomic array, methylation array

Research : Computational biology, systems biology, cancer genomics, tumor microenvironment, epigenetics, data integration, machine learning, biological networks
Methods : Statistical modeling, bioinformatics analysis, high-throughput data analysis, algorithm development, data mining, network inference, predictive modeling
Technologies : Next-generation sequencing, computational pipelines, machine learning frameworks, data visualization tools, high-performance computing, databases, programming languages

Research : Neural stem cells/neurogenesis, niche/microenvironment, cell communication, tissue architecture, stress response, Drosophila model
Methods : Genetic manipulations and labelling, multicolour clonal analysis, live imaging, brain explants, bulk and single-cell omics, behavioural assays
Technologies : Confocal microscopy, Targeted DamID, single-cell RNA sequencing, image analysis, Drosophila genetics

Research : Epigenomics, chromatin biology, gene regulation, DNA methylation, non-coding RNA, developmental biology, genome architecture, transcriptional regulation
Methods : ChIP-seq, ATAC-seq, RNA-seq, DNA methylation profiling, single-cell sequencing, bioinformatics analysis, data integration, functional genomics assays
Technologies : Next-generation sequencing, single-cell sequencing platforms, bioinformatics pipelines, computational biology tools, high-throughput sequencing, microscopy, data visualization software

Research : Chromatin biology, epigenetics, DNA replication, histone dynamics, genome stability, gene regulation, nuclear organization, cell cycle
Methods : Chromatin immunoprecipitation, microscopy, DNA fiber assays, live-cell imaging, molecular biology techniques, protein interaction assays, sequencing
Technologies : Next-generation sequencing, fluorescence microscopy, super-resolution imaging, mass spectrometry, bioinformatics tools, single-molecule analysis, flow cytometry

Research : Epigenomics, chromatin biology, gene regulation, DNA methylation, non-coding RNA, developmental biology, genome architecture, transcriptional regulation
Methods : ChIP-seq, ATAC-seq, RNA-seq, DNA methylation profiling, single-cell sequencing, bioinformatics analysis, data integration, functional genomics assays
Technologies : Next-generation sequencing, single-cell sequencing platforms, bioinformatics pipelines, computational biology tools, high-throughput sequencing, microscopy, data visualization software

Research : Chromatin biology, epigenetics, DNA replication, histone dynamics, genome stability, gene regulation, nuclear organization, cell cycle
Methods : Chromatin immunoprecipitation, microscopy, DNA fiber assays, live-cell imaging, molecular biology techniques, protein interaction assays, sequencing
Technologies : Next-generation sequencing, fluorescence microscopy, super-resolution imaging, mass spectrometry, bioinformatics tools, single-molecule analysis, flow cytometry

Research : Neural stem cells, brain plasticity, adult neurogenesis, stem cell heterogeneity, cell fate decisions, brain homeostasis, regeneration, zebrafish model, spatial organization
Methods : Live imaging, lineage tracing, clonal analysis, genetic labeling, molecular profiling, single-cell omics, spatial mapping, behavioral tracking, time-lapse microscopy, computational analysis
Technologies : Two-photon/confocal microscopy, CRISPR/Cas9, single-cell RNA sequencing, spatial transcriptomics, bioinformatics, image analysis, mathematical modeling, data integration, zebrafish transgenics

Research : Pediatric neuro-oncology, Tumor microenvironment, Tumorigenesis in cerebellar development, Tumor cell plasticity and resistance, Cancer cell heterogeneity, Pediatric cancer immunotherapy, Translational research in childhood oncology
Methods : Proteomics and mass spectrometry, Nanopore sequencing, Multi-omics approaches, ScRNA sequencing, In vivo mouse models, Tumor microenvironment analysis, High-throughput screening for therapeutic targets
Technologies : Advanced sequencing platforms, Mass spectrometry-based proteomics, Single-cell sequencing technologies, Multiplex immunofluorescence imaging, Data integration platforms for multi-omics data, Computational modeling of tumorigenesis

Research : Neural stem cells, brain plasticity, adult neurogenesis, stem cell heterogeneity, cell fate decisions, brain homeostasis, regeneration, zebrafish model, spatial organization
Methods : Live imaging, lineage tracing, clonal analysis, genetic labeling, molecular profiling, single-cell omics, spatial mapping, behavioral tracking, time-lapse microscopy, computational analysis
Technologies : Two-photon/confocal microscopy, CRISPR/Cas9, single-cell RNA sequencing, spatial transcriptomics, bioinformatics, image analysis, mathematical modeling, data integration, zebrafish transgenics
Cell Next | Training, Career Development and Innovation

Research : Chromatin biology, epigenetics, DNA replication, histone dynamics, genome stability, gene regulation, nuclear organization, cell cycle
Methods : Chromatin immunoprecipitation, microscopy, DNA fiber assays, live-cell imaging, molecular biology techniques, protein interaction assays, sequencing
Technologies : Next-generation sequencing, fluorescence microscopy, super-resolution imaging, mass spectrometry, bioinformatics tools, single-molecule analysis, flow cytometry

Research : Developmental biology, gene regulation, cellular differentiation, molecular pathways, stem cells, embryogenesis, genetic networks, signal transduction
Methods : Molecular cloning, gene expression analysis, CRISPR/Cas9 genome editing, microscopy, cell culture, RNA sequencing, functional assays, imaging techniques
Technologies : Next-generation sequencing, fluorescence microscopy, live-cell imaging, bioinformatics tools, flow cytometry, gene editing platforms, laboratory automation

Research : Spatial transcriptomics, computational pathology, high‑content screening, machine learning, phenotypic screening, image‑based phenotyping, weak supervision, annotation reduction
Methods : Deep learning, computer vision, image segmentation, morphological–molecular inference, image simulation, cross‑modality prediction, predictive modeling, data integration.
Technologies : Neural networks, microscopy imaging, spatial transcriptomics, image analysis pipelines, annotation tools, simulation engines, GPU computing, digital slide scanners