Chromatin organisation helps cells to remember their past
Paldi, F., Szalay, M.F., Dufau, S., Di Stefano, M., Reboul, H., Jose, D., Bantignies, F., and Cavalli, G. (2026). Transient histone deacetylase inhibition induces cellular memory of gene expression and 3D genome folding. Nat Genetics, in press. 10.1038/s41588-025-02489-4.
A Cell-ID team led by Giacomo Cavalli discovered that cells are able record information on past exposures in their 3-dimensional (3D) chromatin architecture. Each nucleus contains nearly 2 meters of DNA that is carefully compacted and organised in a way that it supports cellular function. Although such 3-dimensional (3D) genome1 folding collaborates with other epigenetic regulatory mechanisms2 to regulate the expression of the appropriate genes in different cell types, it was not known whether this organisation itself carries critical information beyond the DNA sequence.
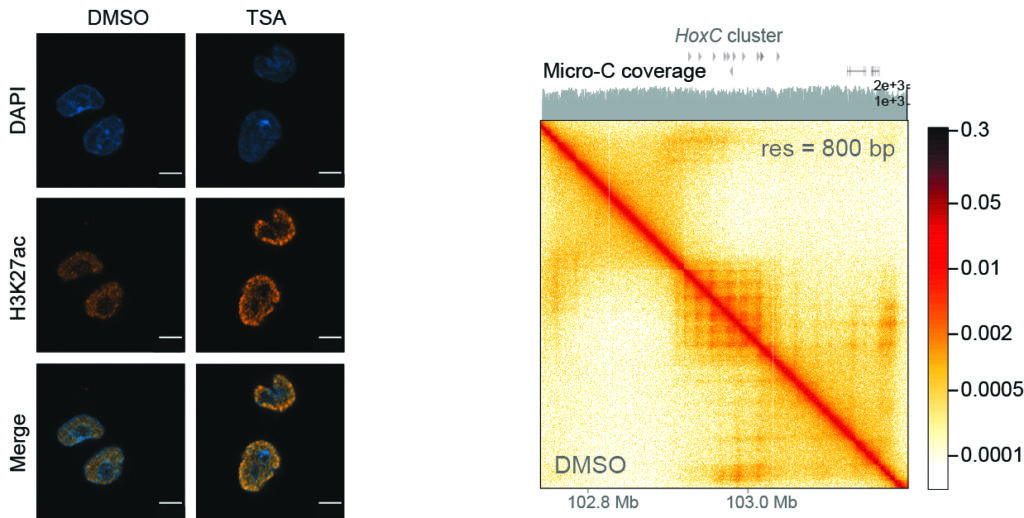
By causing epigenetic dysregulation3 in mouse embryonic stem cells, and then restoring the cells to their normal state, scientists have found that 3D chromatin architecture undergoes deep changes upon an acute cellular challenge. After the end of the challenge and the restoration of the initial conditions, most genes are restored to their normal level of expression and the 3D folding of the genome is also reestablished in the majority of the loci. However, some of the loci partially maintain a perturbed 3D architecture and an altered state of gene expression.
This shows that cells are able to record a transient challenge in the architectural features of their genome. Critically, this allowed cells to respond more robustly to a subsequent perturbation. Furthermore, when certain 3D genome structures4 were experimentally removed, cells could not retain memory of the perturbation. This indicates that chromatin organisation can store critical information on the cells’ past. Such memory process could be important in healthy development as well as disease conditions, where epigenetic perturbators are administered repeatedly to patients.
Notes
1 – The genome is defined as the set of genetic material – and therefore the entire DNA sequence – contained in a cell or organism.
2 – Epigenetics is the study of the mechanisms that allow the inheritance of different gene expression profiles in the presence of the same DNA sequence.
3 – Scientists focused on histone deacetylases, a group of enzymes that negatively regulate the expression of genes by covalently modifying chromatin.
4 – Polycomb proteins are important developmental regulator complexes that partially exert their function by establishing specific chromatin structures.
Reference
Paldi, F., Szalay, M.F., Dufau, S., Di Stefano, M., Reboul, H., Jose, D., Bantignies, F., and Cavalli, G. (2026). Transient histone deacetylase inhibition induces cellular memory of gene expression and 3D genome folding. Nat Genet in press. DOI: 10.1038/s41588-025-02489-4.
Figure copyright: Flora Paldi and Giacomo Cavalli